The Kavraki Lab has introduced an online portal to help researchers screen COVID-19 drug candidates that might attack specific proteins of the SARS-CoV-2 virus.
Lydia Kavraki and her colleagues at the University of Houston, the University of Edinburgh, Scotland, and the Federal University of Ceará, Brazil, have posted a “user-friendly” web server offering scientists the chance to screen their drug candidates virtually in relation to known protein binding pockets on the SARS-CoV-2 virus.
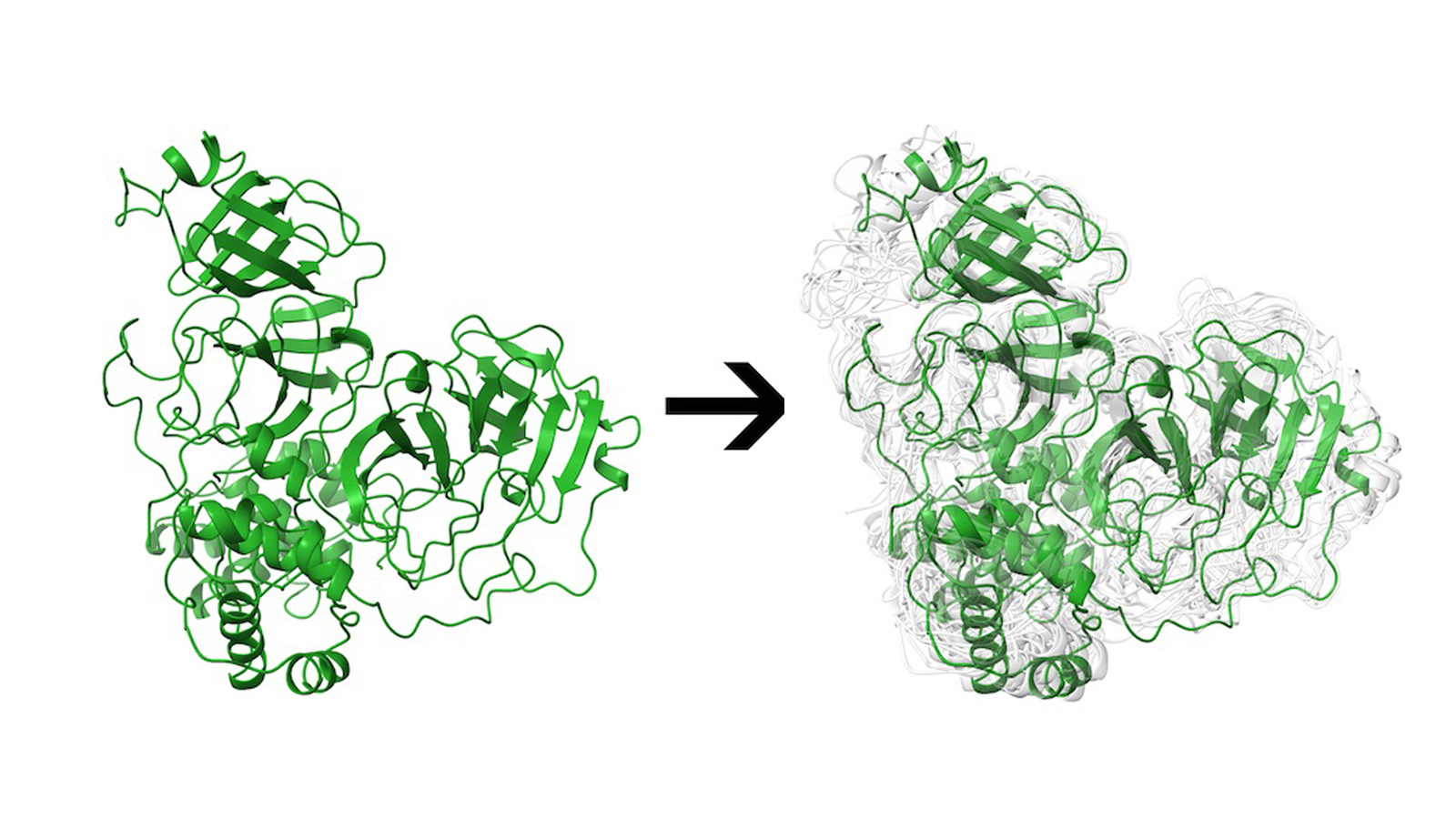
Better yet, the program incorporates what they say is an often-overlooked factor in computational models of these pockets: their flexibility.
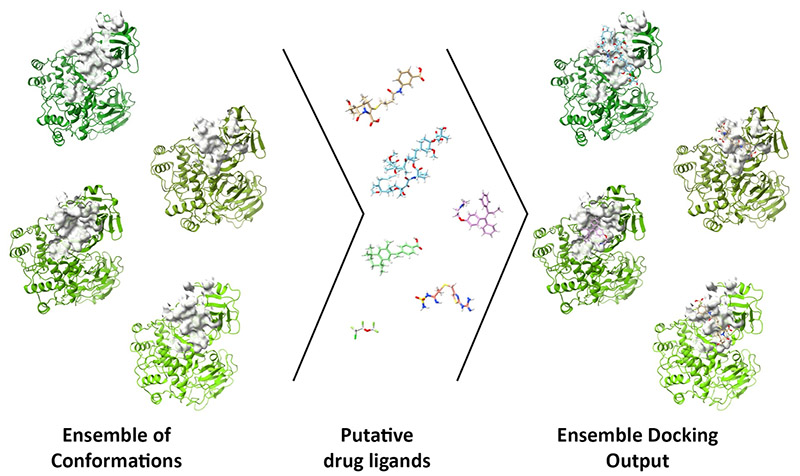
The project, detailed in an open-access paper in Computers in Biology and Medicine, incorporates models of three drug targets—main protease (Mpro), RNA-dependent RNA polymerase (RdRp) and Papain-like protease (PLpro)—for ensemble docking through DINC-COVID.
The ensemble docking approach allows researchers to screen candidate ligands (reactive molecules) against different conformations of SARS-CoV-2 proteins and their binding pockets. DINC-COVID then scores the ligands’ success at binding.
DINC stands for “Docking INCrementally,” a protocol developed by Kavraki’s lab in 2013 to speed protein-peptide docking simulations that help researchers design drugs, vaccines and other processes involving large ligands. An upgraded version led by Kavraki and Dinler Antunes, then a postdoctoral researcher in her lab and now an assistant professor at the University of Houston, appeared in 2017.
The new iteration relies on the “impressive number” of SARS-CoV-2 protein structures that have been resolved so far. Understanding these structures allows researchers to find binding partners that could, ideally, deactivate the virus.
The study also offers a literal twist, best represented by the main protease, a docking site on the virus that has been the focus of much attention over the past 18 months. Researchers have found the Mpro site can substantially distort its shape in response to binding, allowing it to accommodate a diverse set of potential ligands.
That malleability makes Mpro and other sites hard to simulate, with a much higher computational cost, said Rice postdoctoral researcher and co-author Mauricio Rigo. “Unlike other servers, the proteins we are making available aren’t static; they’re not a single conformation,” he said. “We use states to reflect the dynamics of this protein in a physiological environment.”
The team used several programs to narrow the ensembles from the 100,000 possible conformations generated by a molecular dynamics simulation, for instance, to a set of representative conformations. That lets the researchers decouple ensemble generation from docking within DINC-COVID, saving hours or days on complicated calculations.
“We believe this was the right way to go,” Rigo said. “Our tests of the algorithm gave us a good match with experimental results.”
Along with Mpro, the team modeled ensembles of catalytic binding sites on PLpro and RdRp. For Mpro, they modeled its catalytic and allosteric binding sites, for a total of 12 ensembles.
“We chose them because these can be targeted by different drugs,” said Sarah Hall-Swan, a Rice graduate student and co-lead author of the paper. “When you’re trying to find a drug to inhibit a virus, you're going to look for the protein parts that are important for that virus to function and try to inhibit them.”
The lab is working to expand the number of ensembles available in DINC-COVID.
“We are very pleased with the response of the community to our work,” Kavraki said. “DINC-COVID has already been used by about 500 researchers in 16 different countries, while our earlier web server DINC has been accessed by 11,000 users. We hope DINC-COVID will help shed light on the complex mechanisms of infection by SARS-CoV-2."
Didier Devaurs, a former Rice postdoctoral researcher and now a research fellow at the University of Edinburgh, is co-lead author of the paper. Geancarlo Zanatta, an associate professor of physics at the Federal University of Ceará, is a corresponding author. Kavraki is the Noah Harding Professor of Computer Science, a professor of bioengineering, mechanical engineering and electrical and computer engineering and director of the Ken Kennedy Institute.
The National Science Foundation (2033262), the National Council for Scientific and Technological Development of Brazil (437373/2018-5), a University of Edinburgh and Medical Research Council fellowship (MC_UU_00009/2), the Cancer Prevention and Research Institute of Texas (RP170593), the National Institutes of Health (U01CA258512) and a National Library of Medicine Training Program fellowship (T15LM007093-29) supported the research.
Main Image: A Rice University web server can help researchers identify potential drugs to treat COVID-19. In the physiological environment, a protein is not a static entity, but can move and adopt an ensemble of conformations. In this illustration, the main protease of SARS-CoV-2 is represented in green and multiple conformations are represented in light gray. Illustration by Mauricio Rigo/Kavraki Lab