Details about variants hiding in the deluge of genetic SARS-CoV-2 sequences would be good to know, if only researchers can get to them.
A new program developed by the Treangen Lab at Rice University will make it possible, at least for “intrahost variants,” those that appear in genome data from the same COVID-19-positive person.
A Rice team led by CS assistant professor Todd Treangen and graduate student Yunxi Liu has developed Variabel, which accurately identifies “low-frequency variants” of the virus that causes COVID-19. The team's research is detailed in Nature Communications,
"Finding these clues could be key to identifying potentially devastating variants before they have a chance to spread," Treangen said.
The data is freely available, but there’s a lot of it. The research makes low-frequency variant mining available for an estimated half-million SARS-CoV-2 genomes gathered by Oxford Nanopore Technologies (ONT), which offers an affordable platform for rapid sequencing of single, long molecules of DNA or RNA.
“Variabel directly enables the use of affordable nanopore sequencing technology for the identification of within-host variation after viral infection,” said Treangen, whose work has focused on infectious disease monitoring since long before the COVID-19 pandemic.
The lab had similar success in testing Variabel on sequence data from patients infected with Ebola and norovirus.
The open-source program is available for download at https://gitlab.com/treangenlab/variabel.
The researchers claim the key to Variabel is its ability to distinguish true variants from sequencing errors in the ONT process.
To validate Variabel, they compared data taken over time from single positive patients as well as sequences from cross-patient datasets, produced by ONT and another sequencing technique, Illumina. Over time, a single patient can host as many as a billion copies of a virus.
By comparing results before and after applying Variabel to the data, they found the program was able to correct the great majority of sequencing errors.
“Variabel opens the door to portable, affordable and rapid characterization of within-host variation, which ultimately could aid in the discovery of future mutations specific to variants of concern,” said Treangen, whose lab, along with Rice’s Ken Kennedy Institute, hosted a March 11 symposium to discuss scientific advances spurred by the pandemic. The virtual symposium can be viewed online here: http://www.youtube.com/watch?v=YaNm7QBmxD8.
Co-authors of the paper are Rice undergraduate Joshua Kearney and software engineer Bryce Kille, and Baylor College of Medicine postdoctoral associate Medhat Mahmoud and Fritz Sedlazeck, an associate professor at the Human Genome Sequencing Center and an adjunct associate professor in Rice's Department of Computer Science.
The National Institute of Allergy and Infectious Diseases (1U19AI144297, 1P01AI152999-01), a C3.ai Digital Transformation Institute COVID-19 award, the Centers for Disease Control (75D30121C11180), the National Science Foundation (1338099) and Rice’s Center for Research Computing supported the research.
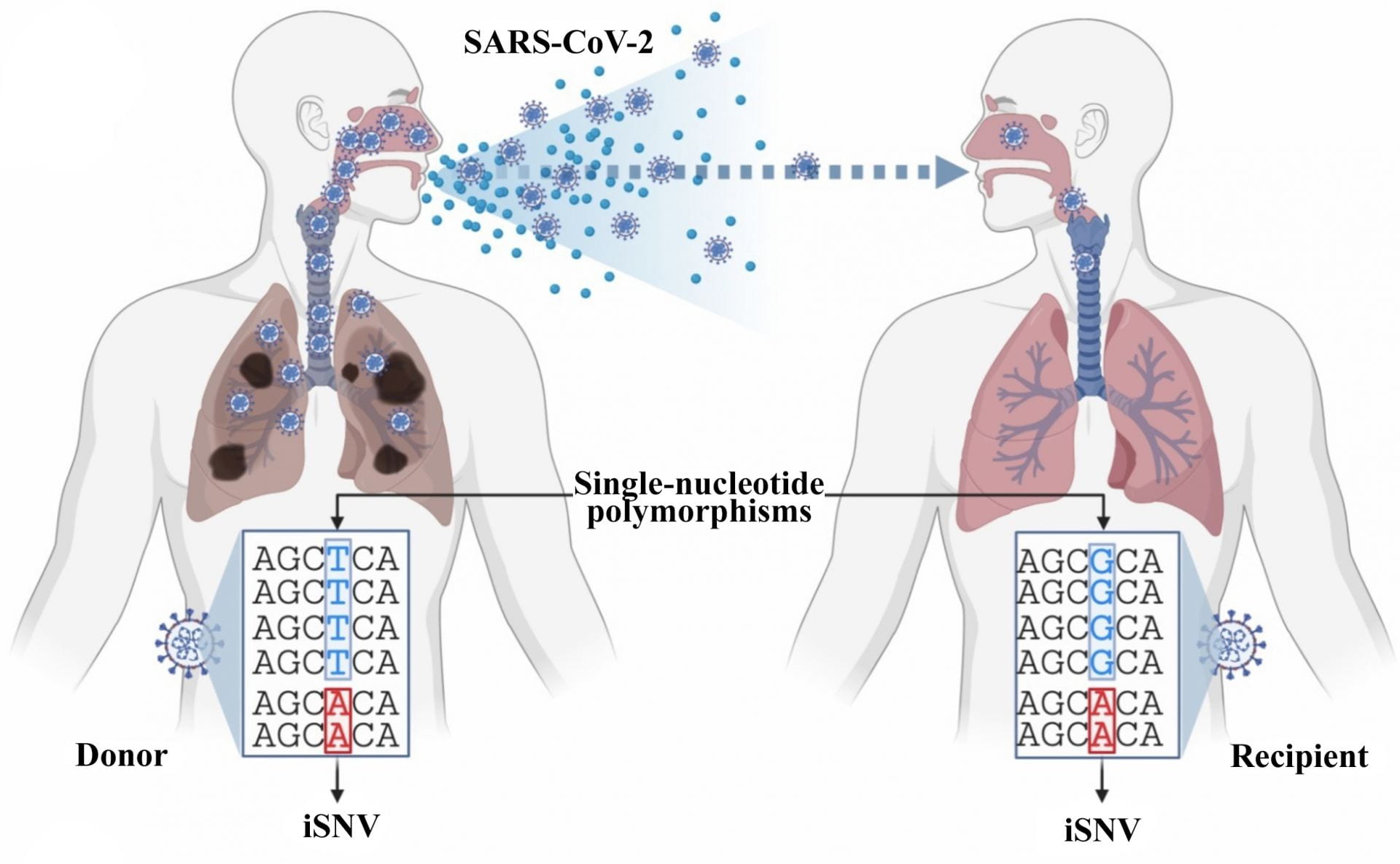
Image Caption: An illustration defines what differentiates single-nucleotide variants (iSNVs) within a single host from single nucleotide polymorphisms that spread from host to host. Rice University computer scientists have introduced Variabel, which uses sequencing data to identify low-frequency, intra-host variants of SARS-CoV-19 from public data sets. Illustration courtesy of the Treangen Lab